
First, you may be asking yourself €“ Why viral bioinformatics? Good question! Although it€™s true that much in the world of bioinformatics can be applied to all manner of protein and DNA sequences, there are a number of resources that are specific for viruses and there are a number of analyses that every virologist should be familiar with. So this section of the virology toolbox will highlight database resources, some useful tools and analyses, and some pitfalls you want to avoid.
What do I know about viral bioinformatics? Another good question! Well, all I can say is that although I can€™t program my way out of a paper bag €“ I€™ve been analyzing DNA and protein sequences of poxvirus for 20+ years and have been developing software for analysis of viral genomes for 10+ years. That is, I say what I want the software to do, and a bunch of talented programmers €“ including many undergraduate students €“ figure out how to code it. Over the years, this work has been funded by NSERC and PENCE in Canada, and NIH in the USA.
I€™ll also be highlighting many of our own tools €“ why?
- They were developed precisely for comparative genomics of viruses. That is, genomes from 10-500 kb.
- They were developed for use by the bench virologist €“ so they€™re fairly straightforward to use.
- They€™re platform independent – will run on Macs, Windoze and LINUX boxes.
- I know them best and can give good examples of their use.
- PS. The programs also work on any type of DNA and protein sequences.
My first topic is very simple, but equally important. So watch your language.
Homology = common origin
Phrases like sequence (structural) homology, high homology, significant homology, or even 35% homology are as common, even in top scientific journals, as they are absurd, considering the above definition.
I took this quote from a book (which seems to be online): Sequence – Evolution €“ Function: Computational Approaches in Comparative Genomics by Eugene V Koonin and Michael Y Galperin: Kluwer Academic; 2003.
So genes/proteins/sequences are either homologous, or they€™re not. No fractions or percentages here!
Try writing: 50% identical. But you also have to say whether you mean nucleotides or amino acids.
While we€™re on this topic:
Orthology
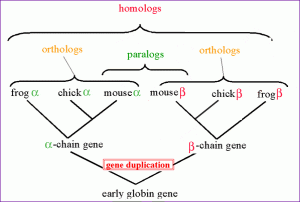
Homologous sequences are orthologous if they were separated by a speciation event: when a species diverges into two separate species, the divergent copies of a single gene in the resulting species are said to be orthologous. Orthologs, or orthologous genes, are genes in different species that are similar to each other because they originated from a common ancestor. The term “ortholog” was coined in 1970 by Walter Fitch.
Paralogy
Homologous sequences are paralogous if they were separated by a gene duplication event: if a gene in an organism is duplicated to occupy two different positions in the same genome, then the two copies are paralogous. A set of sequences that are paralogous are called paralogs of each other. Paralogs typically have the same or similar function, but sometimes do not: due to lack of the original selective pressure upon one copy of the duplicated gene, this copy is free to mutate and acquire new functions (from Wikipedia).
The figure illustrates homologs, orthologs and paralogs (click for the original link).

Comments are closed.