The RNA genome of influenza viruses is segmented . The virions of influenza A and B viruses contain 8 different RNAs, while those of influenza C viruses contain 7. How is the correct number of RNA segments inserted into newly synthesized virus particles?
During influenza virus assembly, viral RNAs and viral proteins €“ called a ribonucleoprotein complex or RNP – travels to the plasma membrane. There the virion forms by a process called budding, during which the membrane bulges from the cell and is eventually pinched off to form a free particle.
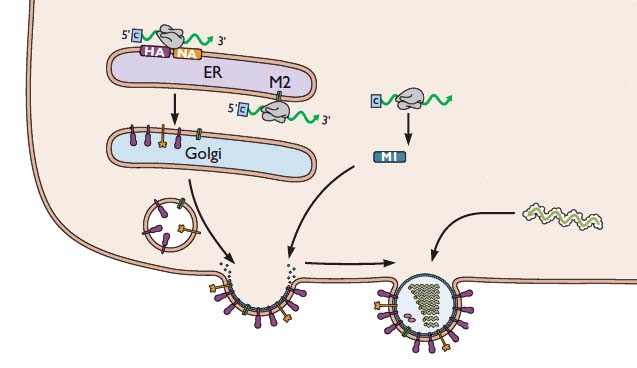

Production of an infectious virus particle requires incorporation of at least one copy of each of the eight RNA segments. Two different mechanisms – random and selective packaging – have been proposed to explain how each virion receives a full complement of genomic RNA.
If the 8 influenza viral RNA segments were randomly packaged into new particles, we would expect to observe 1 infectious particle for every 400 particles assembled (8!/88). This ratio falls within the range of infectious to noninfectious particles that occur in virus stocks. If more than 8 RNA segments could be packaged into each virion, then the fraction of infectious particles would be significantly increased. For example, if 12 RNA molecules could fit into each virion, then 10% of the particles would have the complete viral genome. In support of this mechanism, influenza viruses with more than 8 RNA segments have been observed.
In the selective packaging mechanism, each of the eight genomic RNAs has a different signal that allows incorporation into virus particles. These signals are believed to be within the noncoding and coding sequences at the 5′- and 3′-ends of the viral RNAs. The sequences interact and form structures that are unique to each segment, and which have been shown to be essential for incorporation of each segment into virions. Consistent with this hypothesis, electron microscopy reveals that during budding, the viral RNPs are organized in a distinct pattern, as shown in the image.


This observation argues that RNPs are not randomly incorporated into virions, and is consistent with the presence of specific signals in each RNA segment that enable the RNPs to be packaged as a complete set. The mechanisms by which these signals are recognized, and how they ensure incorporation of one copy of each RNA segment into the particle, are not known.
There is clear evidence for a selective mechanism during the packaging of the bacteriophage ψ6 genome. Viral particles contain one copy each of a S, M, and L dsRNA segment. All particles contain a complete complement of genome segments, as indicated by the fact that every virus particle is infectious. Only the S RNA segment can enter newly formed particles; once that segment is packaged, then the M RNA can enter. The L RNA can only enter particles that contain both the S and M segments. Precise packaging is therefore the result of a serial dependence of packaging of the RNA segments.
Muramoto, Y., Takada, A., Fujii, K., Noda, T., Iwatsuki-Horimoto, K., Watanabe, S., Horimoto, T., Kida, H., & Kawaoka, Y. (2006). Hierarchy among Viral RNA (vRNA) Segments in Their Role in vRNA Incorporation into Influenza A Virions Journal of Virology, 80 (5), 2318-2325 DOI: 10.1128/JVI.80.5.2318-2325.2006
Noda, T., Sagara, H., Yen, A., Takada, A., Kida, H., Cheng, R., & Kawaoka, Y. (2006). Architecture of ribonucleoprotein complexes in influenza A virus particles Nature, 439 (7075), 490-492 DOI: 10.1038/nature04378
Frilander, M. (1995). In Vitro Packaging of the Single-stranded RNA Genomic Precursors of the Segmented Double-stranded RNA Bacteriophage ψ6: The Three Segments Modulate Each Other’s Packaging Efficiency Journal of Molecular Biology, 246 (3), 418-428 DOI: 10.1006/jmbi.1994.0096

how do you determine the ratio of infectious to non-infectious viral particle in a stock?
Why is it that Influenza A,B and C dont mix with one another? Assuming a person could and would be infected with both Influenza A and B, would a recombination happen in this circumstance?
And also, is it the 8th piece of RNA missing in type C that makes this one less pathogenic?
To determine the ratio of non-infectious to infectious particles (a parameter known as the particle-to-plaque forming unit ratio) the infectivity of a virus stock is determined, either by plaque assay or similar technique. Next, the number of physical particles is determined, either by electron microscopy or spectroscopy. Then the ratio of the two number is calculated. Particle-to-pfu ratios vary widely among animal viruses from 1 (meaning every virus particle is infectious) to 10,000 (only 1 in 10,000 virus particles is infectious).
Why influenza subtypes do not reassort is not known. Influenza A reassorts only with influenza A, B only with B, and C only with C. The reason why probably has something to do with the signals on the viral RNAs that specify packaging of the RNAs into the virion. Influenza C viruses have 7 RNA segments, but only because one of these encodes a protein with the function of both the HA and NA (called esterase for influenza C viruses). This protein is called HE (for hemagglutinin-esterase). Why influenza C viruses are less pathogenic than A and B is not known.
Vincent — Here is a recent paper by Perez et al in which ferrets were co-infected with H1 Brisbane (Type A), or H3 Brisbane (Type B) and H1N1 Calif, confirming your point of lack of reassortment.
http://knol.google.com/k/daniel-perez/fitness-o…
But note that the animals coinfected with Type B and H1N1 produced more virus (H1N1) and had more severe symptoms — synergistic interaction, although I don't think Perez used that term.
Now, keeping that in mind, if you look at the CDC timeline for positive samples, say up to Aug22.09, http://www.cdc.gov/flu/weekly/ you see how Type B has been first appearing in December (week 50) and peaking in March (week 9).
Lack of a H1N1/Type B synergism in the spring/summer and the presence of it in the winter results in a very tidy hypothesis to explain the second highly lethal wave of the 1918 pandemic. Of course, the data are with Type B, but lethal synergistic interactions could occur between other respiratory viruses or even bacterial pneumonia and H1N1.
The result would be an H1N1 that looks to virologists as just the same all year in terms of its virulence, but that is presents clinically as more or less dangerous depending on what else is circulating with it.
Forgive this diversion, but I suspect a lot of us are so interested in your discussions because we are trying to get a picture of what is presently going on with the H1N1.
Whoa, sorry. Got my types mixed. H3 is Type A, eh?
But the hypothesis is still plausible. Those CDC data also show H3 positive tests appearing in that week 50 – week15 wave.
I wonder whether co-infection with Type B and H1N1 produces a synergistic effect in terms of symptoms or H1N1 production. . .
but 1918 H1N1 was very virulent alone without synergistic interaction
in lab tests in mice or/and? ferrets
Of course, I came upon this post after my undergraduate lecture on influenza A. This was a question that came up during our discussion: how does the virus package only 8 RNPs and the correct ones? I'll pull from the three papers for next year's lecture and especially look forward to seeing the images in the Noda et al., paper. Thanks, professor.
My pleasure! Keep virology blog in mind for your virology teaching
needs. Always happy to provide help with lecture material.
Hi, Profvrr, how do you know those viruses are non-infectious? Is it possible that they are infectious but don't have a chance to bind to a cell receptor? Is there any motion that will drive a virus to the specific cell receptor?
hey i was wondering if there is any way in which we could monitor the packaging of vial genome?? if not monitor, atleast deduce their interactions and find a mechanism for packing. ?? also is there any way in which v could monitor rna – rna interactions in vivo or invitro ???
– Harini
hey i was wondering if there is any way in which we could monitor the packaging of vial genome?? if not monitor, atleast deduce their interactions and find a mechanism for packing. ?? also is there any way in which v could monitor rna – rna interactions in vivo or invitro ???
– Harini
Pingback: A new type of enveloped virus?
Could it be a combination of both? The virus selects for the minimum required, but some stragglers are able to make it in? Also the selection process is for each segment, but could the segments of each protein could be from different A type viruses that infect the same cell? If so, would this change the RNP and subsequently the transcription mechanism within the virus?
Here is an interesting very recent article – the group uses scanning transmission electron microscopy to examine the structure and arrangement of RNPs in newly budded virions. They present evidence in support of a pseudo-selective model in which RNP length matters, and connections (maybe RNA:RNA interactions) between RNPs may aid proper recruitment of the proper combination of segments.
Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus.Noda T, Sugita Y, Aoyama K, Hirase A, Kawakami E, Miyazawa A, Sagara H, Kawaoka Y.Nat Commun. 2012 Jan 24;3:639. doi: 10.1038/ncomms1647.
What are your thoughts on quanifying particles by qPCR? There are kits available that will detect viral RNA (for example Flu A M1). The complication would be that perhaps some particles may have multiple copies of M1 (and score multiple times) or no copy of M1 (and not score at all). The qPCR result should not distiguish infectious vs. non-infectious.
sorry to join the conversation without getting any invitations. i think you are absolutely right and this is why Professor is talking about the pfu rather than the RNA copies on quantifying the particles.
Thank you very much for this important information