Our discussion of influenza virus replication has so far brought us to the stage of viral RNA synthesis. Last time we discussed the formation of viral RNAs, an event which takes place in the cell nucleus. Now we’ll consider how these RNAs participate in the assembly of new infectious viral particles, as illustrated in the following figure.
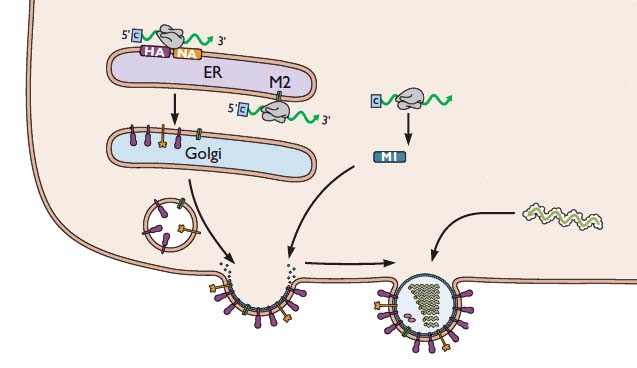

For simplicity, the nucleus is not shown. But remember that the viral RNAs have to be exported from the nucleus to the cytoplasm, where viral assembly occurs. First, the viral mRNAs are translated to produce all the proteins needed to synthesize a new virus particle. The mRNAs encoding the HA and NA glycoproteins are translated by ribosomes that are bound the the endoplasmic reticulum – the membranous organelle that assists in transporting certain proteins to the plasma membrane. As the HA and NA proteins are produced, they are inserted into the membrane of the endoplasmic reticulum as shown. These proteins are then transported to the cell surface via small vesicles that eventually fuse with the plasma membrane. As a result, the HA and NA are inserted in the correct direction in the lipid membrane of the cell. The M2 protein is sent to this location in a similar way.
The (-) strand viral RNAs that will be packaged into new virus particles are produced in the cell nucleus, then exported to the cytoplasm. These RNAs are joined with the viral proteins PA, PB1, PB2, and NP. Viral proteins other than HA, NA, and M2 are produced by translation on free ribosomes, as shown for M1. The latter protein binds to the membrane where HA, NA, and M2 have been inserted. The assembly consisting of viral RNAs and viral proteins – called a ribonucleoprotein complex or RNP – travels to the site of assembly. The virion then forms by a process called budding, during which the membrane bulges from the cell and is eventually pinched off to form a free particle.
As new virions are produced by budding, they would immediately bind to sialic acid receptors on the cell surface, were it not for the action of the viral NA glycoprotein. This enzyme removes sialic acids from the surface of the cell, so that newly formed virions can be released. This requirement explains how the neuraminidase inhibitors Tamiflu and Relenza function: they prevent cleavage of sialic acid from the cell surface. In the presence of these inhibitors, virions bud from the cell surface, but they remain firmly attached. Therefore Tamiflu and Relenza block infection by preventing the spread of newly synthesized virus particles to other cells.

This is fascinating. How do all individual parts “know” when and where to go? How do they find their way in space and time to be at the right location at the precise time?
One way I can explain this is that they are everywhere all the time (well, most of the time). So this means there have to be lots and lots of copies of each individual component everywhere in the cell at a specific time. Is there a mechanism by which the virus “forces” the cell to make its parts instead of the cell's owns (or at least in greater quantities)?
My other question relates to the HA and NA proteins, both found in the virus membrane surface. So, HA binds to the sialic acid, while NA removes it from the cell surface. I'm confused about when the later happens. I'm not sure if it is true, but from the diagrams it appears that HA is 'longer' than NA. So when the virus first attach to the cell through the HA binding to the sialic acid, NA can't reach it, therefore the sialic acid stays. However, during the budding process, NA now is closer to the sialic acid than HA, and now that it's in reach, it can remove it.
How the parts know where to go isn't fully understood. There are
clearly 'signals' on various components that allow them to interact
(protein-protein, or RNA-protein interactions). Proteins destined for
the plasma membrane have specific signals that sort them there. The
cytoplasm is a crowded place, so such signals are very important.
However, viruses often reprogram the cellular biosynthetic activities
so that mainly their proteins/nucleic acids are made. For example,
cell translation is often inhibited so that most of the new proteins
made are those of the virus.
As for NA removing sialic acid – this occurs when the newly
synthesized NA is inserted into the plasma membrane, just before viral
budding occurs. By the time the virion forms, the sialic acid has been
removed. When the virion attaches, it is rapidly taken into the cell
before the NA can work. In cells where the virus is not taken up –
such as red blood cells – the virus will attach, and in time, as the
NA removes the sialic acid, the virus falls off of the cells.
Very informative, interesting on how this virus works. Great article!
Pingback: Packaging of the segmented influenza RNA genome
Pingback: Influenza neuraminidase and H5N1 pathogenicity
the mode of action of tamiflu and relenza is clearly illustrated but it woild have been better if tha mode of action other two vaccine were given.similarly the assembly of virus is slightly unclear so please make it clear.
Hey, I really need help with this project! I am 16 and single guys get at me!
I´m wondering about the function of Neuraminidases, which is only described for the budding process. In the first step of an infectious cycle HA binds to the sialic acid cell receptor. The situation is the same as in the budding process…HA binds to sialic acid. My question is why NA does not cleave sialic acid in the initial step? Or on the other hand are some newly assembled virions again cooperated into the infected cell cause HA should be already active in the newly assembled virions?
I´m wondering about the function of Neuraminidases, which is only described for the budding process. In the first step of an infectious cycle HA binds to the sialic acid cell receptor. The situation is the same as in the budding process…HA binds to sialic acid. My question is why NA does not cleave sialic acid in the initial step? Or on the other hand are some newly assembled virions again cooperated into the infected cell cause HA should be already active in the newly assembled virions?
It is known that ammonia (NH3) can inactivate single-stranded RNA viruses, but the mechanism is not unraveled. I suggest ammonia can twist the RNPs shown down to the right in the picture.
Ottar Stensvold
Ammonia? So thats why those McDonalds burgers were able to kill the viral infections I got from vaccinations!
Maybe the ribonucleoproteins catalyse aminolysis of the RNA?