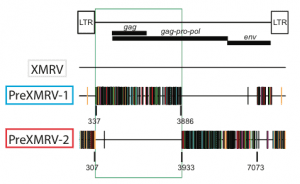

XMRV was originally isolated from a human prostate cancer in 2006, and subsequently associated with ME/CFS. The human cell line 22Rv1, which was established from a human prostate tumor (CWR22), produces infectious XMRV. An important question is whether XMRV was present in the original prostate tumor, or was obtained by passage through nude mice. To answer this question, DNA from various passages of the prostate tumor in nude mice (called xenografts), and the mouse strains used to passage the tumor, were analyzed for the presence of XMRV proviral DNA.
Early-passage xenografts did not contain XMRV, but mouse cells found in them did contain two related proviruses called PreXMRV-1 and PreXMRV-2. The 3€™-3211 nucleotides of PreXMRV-1, and both LTRs, are identical to XMRV save for two nucleotide differences. The genomic 5€™-half of XMRV and PreXMRV-1 differs by 9-10%. PreXMRV-1 is defective for replication due to mutations in genes encoding the gag and pol proteins. PreXMRV-2 does not contain obvious mutations that would prevent the production of infectious viruses. The gag-pro-pol and a part of the env region of this viral genome is identical to that of XMRV save for two base differences; the LTRs and the remainder of the genome differ by 6-12% from XMRV.
Comparison of the sequences of PreXMRV-1 and PreXMRV-2 indicates that recombination between the two viral genomes led to the formation of XMRV. When the sequences of PreXMRV-1 and ˆ’2 are used to construct the recombinant XMRV, the resulting virus differs by only 4 nucleotides from the consensus XMRV sequence derived from all human isolates reported to date.
The nude mice used for passage of the original prostate tumor were likely the NU/NU and Hsd strains. Neither mouse strain contains XMRV proviral DNA, but both contain PreXMRV-1 and PreXMRV-2 proviral DNA.
These data demonstrate that XMRV was not present in the original CWR22 prostate tumor, but arose by recombination of PreXMRV-1 and PreXMRV-2 between 1993-1996. When the original prostate tumor was implanted into nude mice, some of the mice harbored both pre-XMRV-1 and ˆ’2 endogenous proviruses, which recombined to form XMRV. The authors believe that XMRV originating from the CRWR22 xenografts, the22Rv1 cell line, or other related cell lines has contaminated all human samples positive for the virus. In addition, they suggest that PCR assays for XMRV may actually detect PreXMRV-1 and ˆ’2 or other endogenous viral DNA from contaminating mouse DNA.
Another possibility to explain the origin of XMRV is that it arose in mice and can infect humans. If this is true, then XMRV would have to be present in the nude mice used to passage the CWR22 human prostate tumor. No evidence for an XMRV provirus was found in 12 different nude mouse strains, including two used to passage the CWR22 tumor. Furthermore, a screen of 89 inbred and wild mice failed to reveal the presence of proviral XMRV DNA. Hence the authors conclude:
€¦that XMRV arose from a recombination event between two endogenous MLVs that took place around 1993-1996 in a nude mouse carrying the CWR22 PC xenograft, and that all of the XMRV isolates reported to date are descended from this one event.
It is possible that XMRV produced during passage of CWR22 in nude mice subsequently infected humans. Because XMRV arose between 1993-1996, this scenario could not explain cases of prostate cancer and chronic fatigue syndrome that arose prior to that date.
How can these findings be reconciled with the published evidence that sera of ME/CFS patients from the 1980s contain antibodies to XMRV? Those antibodies were not shown to be directed specifically against XMRV, and therefore cannot be used to prove that XMRV circulated in humans prior to 1993-96. Furthermore, in the absence of clear isolation of an infectious virus, antibody tests alone have proven highly unreliable for identification of new viruses.
Where do these findings leave the hypothesis that XMRV is the etiologic agent of prostate cancer and ME/CFS? All published sequences of human XMRV isolates are clearly derived by recombination of PreXMRV-1 and ˆ’2. The finding of human XMRV isolates that are not derived from PreXMRV-1 and ˆ’2 would leave a role for XMRV in human disease. As of this writing, no such XMRV isolates have been reported in the scientific literature.
Update: A second paper has also been published in Science Express today entitled “No evidence of murine-like gammaretroviruses in CFS patients previously identified as XMRV-infected”. Editors of the journal Science have asked the authors to retract their 2009 paper linking XMRV infection with chronic fatigue syndrome. The authors have refused.
T. Paprotka, K. A. Delviks-Frankenberry, O. Cingoz, A. Martinez, H.-J. Kung, C.G. Tepper, W-S Hu, M. J. Fivash, J.M. Coffin, & V.K. Pathak (2011). Recombinant origin of the retrovirus XMRV. Science Express

No, it is impossible for you to conclude that the virus arose in the mid 90s. Not for the rest of us.
What does polytropic and modified polytropic mean? And are the WPI sequences published in the peer reviewed literature?
Two positive studies. 10+ negatives ones. Hmm…. Don’t think I’m really picking and choosing that much, do you? At least I remain open to the possibility that XMRV infects humans, though that’s not how I would place a bet right now.
If you don’t understand host range why do you expect to understand why there is no evidence that the XMRV variant was created in the 1990s?
Only if you find the virus. Â If you don’t, before you publish, you should attempt to replicate. Â Otherwise you cannot claim your results have anything to say about the original paper. Â
Lo et al found polytopic and modified polytropic sequences.  The XMRV variant is a xenotropic and polytropic hybrid.  The WPI also have polytopic and modified polytropic sequences in the GenBank.
You never count studies in science, you use data. Â You could have 10 negative papers on HIV with a new variant and study using a correct assay can find the virus. Â The scientific evidence shows the virus is infecting humans.
No, of course you don’t count studies. You pick* the data you want.
* or would that be “cherry pick..?”
Again, you don’t answer the question.Â
Ms. Whittemore stated, before she even knew about the published data, that the finding that XMRV originated around1993, did not challenge the finding that Lombardi et al. found XMRV in patients that had CFS longe before that  ‘date of XMRV creation’.
Annette Whittemore was provided with the data in advance of publication. Â
You ignore data. Â All the data, every bit of it, says this is a human retrovirus.
Replication is being redefined. Â
So it is logical to assume beforehand that the data in the yet-to-be published paper will be exactly the same as the data that was presented at CROI by Pathak?
Even Ms. Whittemore isn’t as stupid as you imply, and therefore she stated (literal quote by Ms. Whittemore):
“We have not had the chance to review the embargoed Paprotka et al. report”
They had finished the study.
And Ms. Whittemore knew this at the time of her writing because….?
CROI was broadcast on the web.
I would like to see a reply to this also.
dear prof VR…this abstract below is listed as being at cold spring harbour 2011 retroviology meeting…I would be interested if you could explain it to me in more detail and your opinion on it…many thanks for your time…JamesSingh, I.R. “XMRV, or a related virus, is present in a
large percentage of men and is localized to the androgen secreting
Leydig cells of the testis”
You must be living on another planet.
I know. It doesn’t answer the question though.
Pingback: The Neuro Week 5th June 2011 | Health Book
With every single piece of data. Â Â
How does it not? Â Should I be surprised you didn’t say how.
How does a CROI broadcast *prove* that the Science paper does not extend beyond the data presented at the conference? Ms. Whittemore admits that she didn’t know for sure, by the way, when she stated that they had “not had the chance to review” the final Paprotka et al. paper…
It’s really very simple. It is not the question whether the study was finished, but whether it was known that all of the presented data was the data that would be in the Science paper. Thus, your answer “CROI was broadcast on the web” does not answer the question of whether Ms. Whittemore knew of all the data that would be in the Science paper at the time she was writing that letter.
She didn’t, as she admits.
I take it you conclude she is blatantly lying to the editor of Science?
Gerywn or Gob987 or whatever your next handle will be. Just give it up. You are not a scientist with any training, experience nor do you have the educational background to understand the depth of scientific research and analysis. Furthermore, there is no evidence that XMRV even causes ME/CFS.
Realize the fact that if you researched and chronicled the outbreaks of ME/CFS in the last fifty years, you would know that no retrovirus to date, can spread among unrelated patients so rapidly within a 4-5 day incubation period and where I might add is spread via contact and by aerosol especially not from such a half-baked slow replicating virus as XMRV. Please on behalf of the scientific research community, please withdraw back to your flat earth forum.
Guest PA Institute
ERV
I looked at that abstract, it is from Ila Singh’s group, and they use
antibodies to stain tissues obtained from autopsy. The claim is that
mainly prostate stains with the antibodies. However one cannot say
whether the antigens in the prostate are from XMRV or a related
retrovirus. Unfortunately because the tissues are from autopsies one
cannot obtain nucleotide sequence which would be needed to identify a
virus.
To explain this, so everyone understands: the WB is in two parts:
lysates of XMRV-infected cells fractionated in the left panel,
recombinant XMRV gp70 fractionated on the right panel. The positive is
serum from an XMRV infected macaque. Many bands are illuminated
because what is fractionated on this gel is a total cell lysate. The
authors point out what should be p30 and p15, p15E. There may be a
co-migrating species in the lane where CFS serum was used at p30. When
the same serum was used to probe a membrane with gp70, there is no
protein detected. That’s probably why the authors call it a negative.
Even the presence of putative p30 and p15 species in the left panel
does not identify infection with XMRV, just possible cross-reactivity.
As for the PCR – the bands seen in the second round are not the
correct size for a positive, which would be 413 bp.
You know, reading this forum scared me. “In Judy Mikovits I trust”.. Oh my goodness.
They used one assay on one patient and found them positive. Â They then didn’t use that assay on the other patients. Â They used the other assay that had found them negative.
The data was presented at CROI in full.
All patients were first assayed by PCR. One produced bands of
approximately the same size on the second PCR round, so that was
subjected to western blot analysis. The other PCR negative samples
were not examined by western blot. It should be noted that the authors
show all the PCR results, and the western blot of the single specimen.
The reviewers agreed that these constitute a negative finding. It’s
possible that this one patient could be positive for a xenotropic
murine leukemia virus – but if it’s XMRV then it is likely a lab
contaminant. Other murine gamma retroviruses might infect people, and
it is worth finding out if they do.
Is it ok then for Levy to have used the least sensitive PCR in Lombardi et al. Reduced the DNA concentration and raise the annealing time and temperatures to make the assay even less sensitive and compare it to the most sensitive test in Lombardi et al? Then be shocked because his unvalidated assay could not detect a HGRV when the more sensitive assay used by VIP-Dx could?
Do you deny that that this could be an alternative explanation for the failure to detect HGRVs in ME/CFS patients found to be positive by VIP-Dx using methods which detected gag sequences in 67%, but only 4% of controls?
You are a grad student with a consistant ability to be wrong. Good luck with that.
Yet, Lombardi et al have an assay that is specific to MLVs. All of them but mouse ERVs.Â
Please Professor deny that human to mouse transfer of xmrv is possible.
 Yup, only a grad student. Oh wait… he or she has probably still had considerably more training than you have. Who should I trust? Also, grad students can (and should be) compared to “scientists” (which you call PhD-holding researchers, I’m assuming). Grad students have to publish high-quality papers even to finish grad school. Papers that compete with everything that “scientists” try to publish as well.
As an aside, your comments are entirely asinine, Gob. Grow a few brain cells and get back to me.
Pingback: Murine gammaretroviruses in prostate cancer cell lines
>> How can these findings be reconciled with the published evidence that
sera of ME/CFS patients from the 1980s contain antibodies to XMRV?
>> These data demonstrate that XMRV was not present in the original CWR22
prostate tumor, but arose by recombination of PreXMRV-1 and PreXMRV-2
It may happen that the XMRV exists from a lot of time ago, and that the process of converting PreXMRV-1 and 2 as XMRV is something likely to happen assiduously, so when scientists saw it in lab was one of the a lot of times it happens more than expected. Probably the recombination is something that happens daily and the lab got a shot, by chance.
Pingback: Trust science, not scientists
Pingback: Cleaning up after XMRV
I just could not depart your website prior to suggesting that I really enjoyed the standard information a person provide for your visitors? Is going to be back often to check up on new posts