Starting with a bacterial protein, directed evolution in the laboratory has been used to produce a virus-like capsid that binds and protects RNA. This finding has implications for the origins of viruses.
One view of the evolution of life is that viruses were present even before the first cells in the form of self-replicating molecules. These were technically not viruses, because they did not need cells for their replication. The idea is that when cells did arise, the replicators invaded them and then recruited host proteins for the formation of capsids.
The starting point for experiments intended to provide insight into how a protein might become a viral capsid is a bacterial protein, lumazine synthase, that forms 60-subunit particles that have no affinity for nucleic acids. The protein was redesigned and linked to a peptide that binds to an RNA stem-loop called BoxB. It was then further modified for the ability to package an mRNA that encodes the synthase. Packaging was stimulated by adding BoxB tags to the ends of the mRNA. However, only one of 8 particles packaged the mRNA.
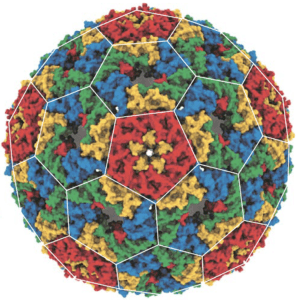
To improve the capsid, the bacterial gene was mutagenized and capsids were selected by rounds of incubation with ribonuclease of smaller and smaller size. The idea was that this strategy would select for capsids that not only packaged the mRNA but would protect it from RNAse digestion. The result was a protein that could form a capsid called NC-4 (pictured) which encapsidated mainly the mRNA and protected it from digestion.
The changes that occurred during this evolution process led to the production of what looks very much like a viral capsid. It is composed of 240 protein subunits arranged in pentamers and hexamers with t=4 icosahedral symmetry*. The amino acid changes that led to the formation of this capsid can be readily discerned. The pores on the capsid are very small, explaining its relative nuclease resistance compared with earlier versions. In addition, the RNA appears to play a role in assembly of the capsid. Finally, changes in the mRNA appear to have led to formation of a stem-loop which facilitates packaging of the nucleic acid into the particle.
These results are amazing: we know that arranging proteins with icosahedral symmetry is an efficient way to build a stable virus particle with the least number of subunits, confirmed by the directed evolution of an icosahedrally ordered and stable capsid.
These observations have multiple implications. They show how ancient self-replicating RNA molecules, the precursors of viruses, might have recruited a host protein and driven its evolution into a protective capsid that specifically packages only the viral RNA. They also indicate how stable particles might be designed as alternatives (dare I say improvements?) to viruses for therapeutic purposes, such as gene therapy and vaccination.
*If you do not understand what icosahedral symmetry is or what t=4 means, please watch my lecture on virus structure.

Pingback: Evolution of a bacterial protein into a virus-like, RNA binding capsid - Virology Hub