Many virology terms are being used these days by people who do not understand their meaning. Included are journalists, medical doctors, scientists, lawyers, and people from all walks of life. In normal times this word mis-usage would be so rare that it would not matter. However, because we are in a viral pandemic that affects nearly everyone, I will attempt to explain the meaning of virus isolates, variants, and strains.
Many of the terms used in virology are ill-defined. They have no universally accepted definitions and there is no ‘bible’ with the correct meanings. As each of us are trained by other virologists, we hear them using terms in certain contexts and we copy their usage – whether or not it is correct. I learned many good things from my mentors but also many things that are wrong.
Nevertheless, certain terms should have specific meanings. Some of my colleagues will certainly disagree with some of my definitions, others will agree. Kudos to the latter. I also recognize that few will read this post and it will have little impact. Perhaps one day a high school student will search for some of the terms and come across it. It is mainly meant for me to put my thoughts down in an orderly manner.
The virology terms I have in mind all have to do with attempts to place order on the huge varieties of viruses in the virosphere. Most of them today derive their meaning from the viral genome: the DNA or RNA that encodes the production of new virus particles. This reliance on the genome is relatively recent: until the 1980s we had no genome sequences; hence most categories were based on other properties, such as the size of the virus particle, whether or not it has a membrane, its type of symmetry, and much more. Today it’s all about the genome. Whether or not you think this myopia is a good idea is not the topic of this post.
Let’s start with the term virus isolate, because it’s the easiest to define. An isolate is the name for a virus that we have isolated from an infected host and propagated in culture. The first isolates of SARS-CoV-2 were obtained from patients with pnemonia in Wuhan in late 2019. A small amount of fluid was inserted into their lungs, withdrawn, and placed on cells in culture. The virus in the fluid reproduced in the cells and voila, we had the first isolates of the virus.
Virus isolate is a very basic term that implies nothing except that the virus was isolated from an infected host. An isolate comes from a single host. We can have my virus isolate, or yours, or the neighbor’s down the street. Most patients do not get to have virus isolates taken from them. Even though SARS-CoV-2 has infected millions, we do not have millions of isolates, probably just thousands. We do have genome sequences from many people, and those can be inferred to represent the isolate from each person – however in most cases infectious virus is not isolated from individual patients.
Isolates are given names so that their origin is known. For example, one of the early isolates of SARS-CoV-2 is called BetaCoV/Wuhan/WIV04/2019. This isolate name consists of the genus, Betacoronavirus, followed by the city of origin, the isolate number, and the year. SARS-CoV-2 is the name of the virus; it is not an isolate name. Isolates of other viruses are also precisely named. I’m a big fan of the very detailed influenza virus nomenclature, which is as follows: Virus name/antigenic type/host of origin if other than human/geographical origin/serial number/last two digits (or all four digits) of year of isolation/hemagglutinin subtype neuraminidase subtype. Examples include influenza A virus A/duck/Germany/1868/68 (H6N1) or influenza A virus A/chicken/Vietnam/NCVD- 404/2010 (H5N1).
A virus variant is an isolate whose genome sequence differs from that of a reference virus. No inference is made about whether the change in genome sequence causes any change in the phenotype of the virus. The meaning of variant has become clouded in the era of whole viral genome sequencing, because nearly every isolate may have a slightly different genome sequence. Such is the case for SARS-CoV-2: nearly every sequence from a different person is slightly different. Up until the end of 2020, any SARS-CoV-2 sequences from any two individuals differed by about ten nucleotide changes out of 30,000. They are all variants, but the term is rarely used in this context. However since then viral genomes with many more changes have been identified. These have been called ‘variants of concern’ (VOC) because it is thought that the changes confer new phenotypic properties such as increased fitness. British scientists did a good deed by calling them VOCs, because now the press must call them variants.
Unfortunately mainstream media, following in the footsteps of scientists who really should know better, have been using the term ‘strain’ to describe what are actually variants. This practice emerges in every viral outbreak: there is a new, more (fill in the blank with your favorite phenotype) strain of Ebolavirus, of Zika virus, and now of SARS-CoV-2. It began early in 2020 with the finding of variants with a single amino acid change in the spike protein, from D to G at position 614. The press called this a new strain that was more transmissible. But the use of strain was incorrect: it is a variant and remains so to this day.
A virus strain is a variant that possesses unique and stable phenotypic characteristics. Such characteristics can only be ascertained by the results of experiments done in the laboratory, in cells in culture and in animals, coupled with observations made in infected humans. The name strain is not easily earned: certainly it cannot simply be given by journalists! As Jens Kuhn has written, €œThe designation of a virus variant as a strain would be the responsibility of international expert groups€. No such designation of strain has been given more than once to SARS-CoV-2: there is one, and only one strain of this virus. No incorrect usage of that term will change this fact. As you might imagine, it can take some time for an international group of experts to agree on anything.
Viral strains are few and far between: it is a designation highly desired but given sparingly. A retrovirologist recently assured me that there is only one strain of HIV-1. The Lansing strain of poliovirus is derived from a human isolate that was passaged 99 times in mice until it acquired the ability to infect that species. That strain has demonstrably different properties from the human strain.
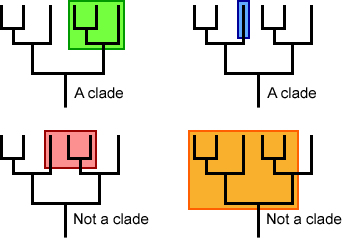
There are other terms to describe viruses but they are more confusing than contentious, and they are not used universally. The term serotype is used to describe viruses of the same species that are antigenically different. There are three serotypes of poliovirus; if you are infected with type 1, then immunity you generate will not protect you against infection with types 2 or 3. Same for the four serotypes of dengue virus, and the hundreds of rhinovirus serotypes. These days, the genome sequence of the virus is used to infer whether isolates are serologically different. The term genotype is used to describe the genetic makeup of a virus. For example, hepatitis C viruses are placed in different genotypes depending on the overall identity of their genomes. For other viruses, the term clade is used. A clade is a group of organisms composed of an ancestor and its descendants, as illustrated by the phylogenetic tree below. SARS-CoV-2 isolates and HIV-1 isolates are placed in clades based on phylogenetic trees constructed from their genome sequences.
I believe that the terms of virology should be used accurately and consistently. The terms isolate, strain, and variant have been frequently and incorrectly misused during the pandemic, which generates confusion. I have little faith that either the general public or the scientists will agree on any nomenclature. Rest assured that if you misuse isolate, variant, or strain, I will correct you according to my lexicon.

Comments are closed.