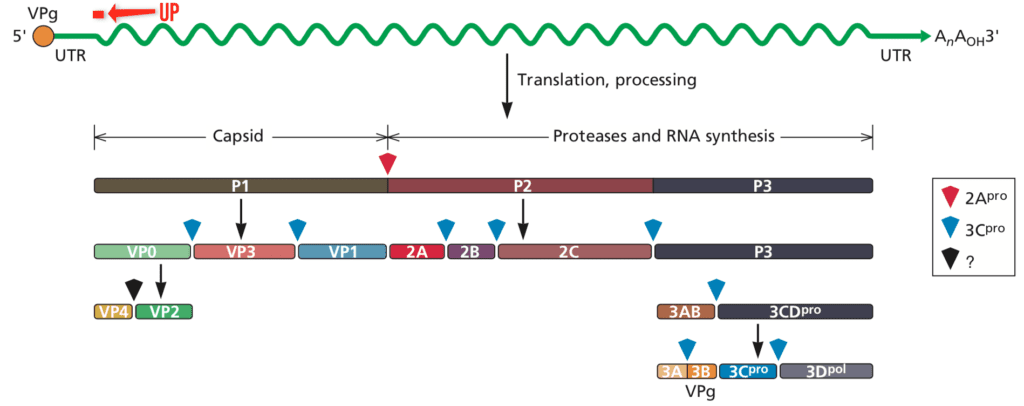
For over 50 years the RNA genome of picornaviruses (illustrated below for poliovirus) was thought to be translated into a single, long polyprotein. All this time a very small upstream open reading frame (uORF) has gone undetected – until now.
Analysis of over 3000 picornavirus genomes revealed a small ORF, beginning with an AUG codon, in the 5-untranslated region of the genome. This uORF (pictured above as a red line) is in a different reading frame from that of the polyprotein. The putative protein, called UP, could be detected in cells infected with the picornaviruses echovirus 7, poliovirus 1, and enterovirus A71.
UP is required for efficient replication in intestinal epithelial cells but not in laboratory cell lines. The protein contains a transmembrane domain, and is associated with the endoplasmic reticulum. A function of UP is to disrupt cell membranes and allow the release of virus particles entrapped in vesicles.
Curiously, UP is only found in the genomes of some enteroviruses, and not in rhinoviruses or other picornaviruses that cause respiratory infections. The protein might be specifically required for replication in intestinal epithelial cells.
When I saw this manuscript a few days ago, it reminded me of work we had done in the 1980s. We had just completed the nucleotide sequence of the poliovirus genome and found clear evidence for a single polyprotein initiated by an AUG codon at nucleotide 742. We were surprised to find 7 other AUG codons in the putative 5€™-noncoding region, and were interested in determining whether these were initiators of protein synthesis.
We mutated each of the 7 AUG codons in the poliovirus genome to UUG and found that 6 of the 7 viruses replicated normally. The exception was a virus with a mutation of AUG #7: this virus formed small plaques on HeLa cells and replicated poorly. We surmised that this mutation somehow affected translation of the polyprotein.
Why didn€™t we see the UP uORF back in 1988? Our experiments were done with type 2 poliovirus, not type 1. The former has a very short open reading frame after AUG #7 and consequently we did not think it was important. AUG #7 of poliovirus type 1, in contrast, has a longer reading frame, encoding UP, that overlaps the polyprotein reading frame (see illustration below).


We were studying type 2 poliovirus because it could paralyze mice, allowing us to study the determinants of neurovirulence. Had we used type 1 poliovirus, we might have seen the UP uORF and detected its encoded protein back in 1988.

Pingback: The lost picornavirus ORF – Virology
Pingback: The lost picornavirus ORF -