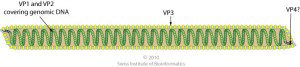

Thermoproteus tenax virus 1 (TTV1) infects the hyperthemophilic archaeon Thermoproteus tenax, which grows at 86°C. The enveloped virus particles are flexible filaments 400 nm long and 40 nm in diameter (illustrated; image credit) built with four capsid proteins, TP1-TP4. The basic proteins TP1 and TP2 bind the 16 kb double-stranded DNA genome to form the nucleocapsid.
Thirty years after the discovery of TTV1, the capsid proteins remained ORFans – meaning that they had no sequence homology with viral or cellular proteins. Recently a more sensitive homology analysis revealed that TP1 is similar to Cas4, a nuclease that is a part of the prokaryotic CRISPR-Cas defense system.
Although TP1 clearly matches the Cas4 protein, it is not complete: codons at the carboxy-terminus are missing. A re-examination of the TTV1 genome sequence revealed a previously undetected open reading frame of 74 codons just downstream of the TP1 gene which are the missing C-terminal residues of the Cas4 nuclease. It is not known if this protein, called gp7, is produced in infected cells; it is not part of the virus particle.
Together the TP1 and gp7 proteins represent a full length Cas4 nuclease. TP1 is probably not catalytically active due to amino acid changes in the active site of the enzyme.
Why does TP1 lack the carboxy-terminal residues of Cas4? The amino terminus of the TP1 protein comprises a positively charged surface that might be involved in binding the viral DNA genome. The same surface in Cas4 is covered by the carboxy-terminal domain of the protein. This observation suggests that transformation of Cas4 from a nuclease into a viral capsid protein probably required removal of this shielding domain, so that the protein could bind the DNA genome.
How did a nuclease become a viral capsid protein? An ancestor of TTV1 might have encoded a Cas4-like protein with nuclease activity with a role in genome replication or repair. Mutations causing loss of nuclease activity might have been followed by truncation of the protein to expose the DNA binding domain, which then became a viral capsid protein. Support for this idea comes from the observation that a Cas4-like protein encoded in the genome of another archaeal virus, the rudivirus SIRV2, has nuclease activity.
Exaptation, a change in the function of a protein during evolution, is known to have taken place in the viral world. The case of Cas4 and TP1 shows that capsid components can evolve from proteins with a very different function.

Pingback: Exaptation: A cell enzyme becomes a viral capsi...
Pingback: TWiM 120: Snakes in trouble | This Week in Microbiology