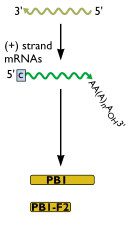

Let’s resume our discussion of the influenza virus genome. Last time we established that there are eight negative-stranded RNAs within the influenza virion, each coding for one or two proteins. Now we’ll consider how proteins are made from these RNAs.
Figure 1 shows influenza RNA segment 2, which encodes two proteins: PB1 and PB1-F2. The (-) strand viral RNA is copied to form a (+) strand mRNA, which in turn is used as a template for protein synthesis. Figure 2 (below) shows the nucleotide sequence of the first 180 bases of this mRNA.
The top line, mostly in small letters, is the nucleotide sequence of the viral mRNA. During translation this sequence is read in triplets, each of which specifies an amino acid (the one-letter code for amino acids is used here). Translation usually begins with an ATG which specifies the amino acid methionine; the next triplet, gat, specifies aspartic acid, and so on. Only the first 60 amino acids of the PB1 protein are shown; the protein contains a total of 758 amino acids.
Most of the influenza viral RNAs code for only one protein. However, RNA 2 (and two other RNAs) code for two proteins. In the case of RNA 2, the second protein is made by translation of what is known as an overlapping reading frame.
On the second line of the RNA sequence in figure 2 is an atg highlighted in red. You can see that this atg is not in the reading frame of the PB1 protein. However, it is the start codon for the second protein encoded in RNA 2, the PB1-F2 protein (F2 stands for frame 2, because the protein is translated from the second open reading frame). Figure 3 shows how PB1-F2 is translated. The sequence of the viral RNA is shown from the beginning, except that reading frame 1, which begins at the first ATG, is not translated. Rather, we have begun translation with the internal atg, which is in the second reading frame. This open reading frame encodes the PB1-F2 protein which, in this case, is 90 amino acids in length (its length varies in different isolates). The protein is much shorter than PB1 because translation stops at a termination codon (tga) long before the end of the RNA. Because PB1-F2 is encoded in reading frame 2, its amino acid sequence is completely different from that of PB1.




The sequences used for this example are from the 1918 H1N1 strain of influenza. Notice the amino acid of PB1-F2 which is highlighted in blue. This amino acid has an important role in the biological function of the protein, which we will consider in a future post.
My apologies if the figures and text are not optimally aligned. A blog post is not the optimal format for such information, but in the interest of time I have not explored other options. Suggestions for improvement are welcome.
Send your questions to virology@virology.ws.

1) So the second PB1-F2 protein (@ 90 amino acids) is tiny compared to the overarching PB1 protein?
2) The PB1-F2 internal atg tag starts at frame 32. But there is an atg at frame 16. So all atg sequences must not be start codons for new protein sequences?
Just re-read. And I don't think I even used word “frame” correctly. Let me try again:
2) The PB1-F2 internal atg tag starts at amino acid triplet 32. But there is an atg at triplet 16. So all atg sequences must not be starter codons for new protein sequences?
That's right, not all atg codons are used to initiate translation.
And, the PB1 protein is much larger than PB1-F2.
Thanks so much.
As someone with an undergraduate background in mathematical linguistics, you can imagine I find this material on multiple coding fascinating. I actually did some original work (building on the second order predicate calculus of Russell and Whitehead in Principia Mathematica) in third order predicate calculus, working on a metalanguage to encode varying levels of ambiguity into machine readable form.
That work directly addressed the artificial intelligence problem of why do computers have such a hard time providing meaning to the phrase:
“Time flies like a butterfly, but fruit flies like a banana”.
That phrase may sound like nonsense, but my work work dealt with how you code the ambiguity immanent in such phrasing so as increase redundancy (and therefore understanding).
And in my mind, the way this particular RNA2 codes this is formally a deeply similar coding issue.
I'm making my way through the material and do not understand it all, which is to be expected. I'm trying to study it further and will try to ask competent, non-trivial questions.
Pingback: Poliovirus @ Operation Willi
This is probably a silly question , but shouldn't those T's be U's, since we are dealing with RNA and not DNA here? For instance, the startcodon in mRNA would be AUG instead of ATG, would it not?
Leti, Yes you are right. It should be AUG which codes for Met.
Yes, we are discussing RNA here. But no one determines sequences of
RNA any longer; the RNA is converted to DNA, and the sequence of the
DNA is determined. Therefore all the sequences in public databases are
given as DNA, not RNA. Those who work with such sequences understand
that many of them represent RNA, and do not spend time converting the
sequence to the RNA bases.
Would you please comment on this.I have done approx. 5000 hours of study on the entire spectrum of pandemic-related issues over the last 12 years.I have two videos on You Tube on the topic.Over 50 years ago,I kept hearing something telling me “the answer is in lemons”.And now…with the H1N1 “dating” the H5N1 virus in various mammals …we face the probability of something nasty down the road.I have been shocked to learn in recent years of the ability of various lemon-based compounds to thwart adhesion of viral particles in the lining of the upper and lower respiratory tract.Could you share with me any serious studies of the possibility of delivering atomized lemon as an attempt to stop/diminish viral load?? Thank you. Could you also answer by email at spiritgift@rocketmail.com ?? Thx.
Plants are known to contain a variety of antimicrobial compounds, but
at concentrations too low to be therapeutically useful. Hence it is
necessary to either purify the compounds or synthesize them; atomized
lemon would not be effective. Several studies have shown the presence
of inhibitors of influenza virus in citrus. For example: “CYSTUS052, a
polyphenol-rich plant extract, exerts anti-influenza virus activity in
mice” was published in Antiviral Research, October 2007, pages 1-10.
In this study, the mice were given an extract of Cistus incanus by
aerosol. In another example, a derivative of hesperidin, a flavonoid
obtained from citrus fruits, was synthesized and shown to inhibit
influenza viral replication. See Biol Pharm Bull. 2009
Jul;32(7):1188-92.
Your blog is very interesting. I would like to tell that I have been looking for such information and finally got it. Thanks a lot.
regards
charcoal grill
In the 2nd paragraph you said that segment 2 of the Influenza genome encodes for the proteins PB1 and PB1-F2, but in the genome section of the website it is shown that genome segment 1 encodes for these proteins, which one is correct? Also when you mapped out the genome segment above, I noticed you had Thymines in there. I was under the impression that thymine was a nucleotide found only in DNA and that once transcribed to RNA the thymine was converted to Uracil? Just wondering if it is a typo or if somehow DNA can can be translated.
Good catch on the PB1, PB1-F2 error. These proteins are encoded by RNA
2. The genome page
(https://virology.ws/2009/05/01/influenza-vir…) has an
older image which is clearly wrong. I'll replace that today. With
respect to T in the sequence, it is not a typo. Since virtually all
nucleotide sequences, even of RNA viruses, are determined by
sequencing a DNA copy, the T is used because that is what is
determined. DNA cannot be translated.
Good catch on the PB1, PB1-F2 error. These proteins are encoded by RNA
2. The genome page
(https://virology.ws/2009/05/01/influenza-vir…) has an
older image which is clearly wrong. I'll replace that today. With
respect to T in the sequence, it is not a typo. Since virtually all
nucleotide sequences, even of RNA viruses, are determined by
sequencing a DNA copy, the T is used because that is what is
determined. DNA cannot be translated.
Be a good listener. Remember, dating you want to get to know the person
There is this crazy guy talking of influenza haplotypes any one ever heard of this
it is very nice post…
jake wand
i find your blog very interesting lots of good
post and readable articles. Keep it up i will surely bookmark your site and
visit it for future readings!
I am really not too familiar with this subject
but I do like to visit blogs for layout ideas. You really expanded upon a
subject that I usually don’t care much about and made it very exciting. This is
a unique blog that I will take note of. I already bookmarked it for future
reference. Thank you..
Now these days influenza is more dangerous, Please share more info about that
If the RNA is transcribed from this originating DNA it will be made in reverse of this and it would begin at the end. Why is the DNA sequence not the complement to the RNA that is made? Why does the convention convert the U’s to T’s only? Thanks.
If the RNA is transcribed from this originating DNA it will be made in reverse of this and it would begin at the end. Why is the DNA sequence not the complement to the RNA that is made? Why does the convention convert the U’s to T’s only? Thanks.
This is the perfect blog for anyone who wants to know about this topic. You know so much its almost hard to argue with you  Housefull 2 Songs
nice article love to hear more on this topic
This doesn’t make as much sense to me as it should. How could a coincidence so perfect happen? I’m probably not understanding this perfectly, but it seems to me that the idea of coding two proteins into a single segment like this is similar to the idea of coding two stories into a single book by moving every space one character to the right.
Dog bites man would become: Dogb itesm an.
I Like the way which you use to structure the article. In like way you put unmistakably a bewildering data in this subject. Appreciative concerning allowing such not to an uncommon degree astounding data to us! Continue Posting. Article Writing