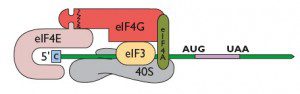

A recent report in The EMBO Journal has revealed a novel modification of the cellular translation apparatus in cells infected with Sin Nombre virus, a hantavirus.The authors show that the viral nucleocapsid (N) protein binds with high affinity to the cap structure on cellular mRNAs. The N protein can also bind the 43S preinitiation complex (which consists of the 40S ribosomal subunit, several initiation proteins, and the met-tRNAi). Finally, N protein has RNA helicase activity, which facilitates ribosome movement through areas of RNA secondary structure. This viral protein therefore functionally replaces all three components of eIF4F: eIF4E (the cap-binding protein), eIF4G (the scaffolding protein which connects the ribosome to the mRNA), and eIF4A, an RNA helicase. It does so even though it has no amino acid similarity to the proteins of eIF4F. Furthermore, the N protein was previously shown to be involved in viral RNA replication and encapsidation. The multifunctional nature of the N protein should come as no surprise: the hantavirus genome encodes only four proteins. Each must therefore fulfill multiple functions in the replication cycle.
Why would the hantavirus genome encode a protein that replaces eIF4F? One of the earliest cellular responses to virus infection is inhibition of translation;the goal is to restrict viral spread. The properties of the N protein could enable unabated viral translation in the face of such a cellular defense. Furthermore, many viral genomes encode proteins that inhibit viral translation. No such activity has been described in cells infected with hantaviruses. Nevertheless, the N protein could permit translation of viral mRNAs when that of cellular mRNAs is inhibited.
The participation of the hantavirus N proteins in multiple events in the cell identify it as an excellent target for therapeutic intervention.
Mohammad A Mir, Antonito T Panganiban (2008). A protein that replaces the entire cellular eIF4F complex The EMBO Journal, 27 (23), 3129-3139 DOI: 10.1038/emboj.2008.228

Hi Vincent – just discovered your blog: looks great! And thanks for pointing out that article on the hantavirus N protein – somehow I'd missed it. Hope you remember me from my time in Jim Hogle's lab (has he entered the blogosphere?). I have recently started myself at http://network.nature.com/people/scurry/blog. Best, Stephen Curry
Cheers – will do!
Hi Stephen – of course I remember you! Thanks for the link to your
blog – I'm now following the RSS feed.
Jim has not entered the blogosphere – I've noticed that there is a
high energy barrier for older scientists (although I certainly am an
exception). One day I'll get him on TWiV, though, and there will be no
turning back.
I must say that blogging about viruses has got me thinking about the
field more broadly than I ever have before. It can only be good!
Stay in touch, and don't hesitate to comment or correct!
Vincent
Hi Vincent – just discovered your blog: looks great! And thanks for pointing out that article on the hantavirus N protein – somehow I'd missed it. Hope you remember me from my time in Jim Hogle's lab (has he entered the blogosphere?). I have recently started myself at http://network.nature.com/people/scurry/blog. Best, Stephen Curry
Cheers – will do!
Pingback: The hantavirus N protein strikes again