
Mycoplasma is a genus of bacteria that are the smallest known free-living organisms. They also have the smallest genomes of any autonomously replicating cell: the DNA of Mycoplasma genitalium is just over one million base pairs in length and encodes 525 genes.
The entire 1,078,809 bp genome of Mycoplasma mycoses was synthesized in 2010 and transplanted into cells of another species, where it replaced the resident genome. In another nod to computer science, the authors refer to ‘installing’ the new genome into a cell, much like a new OS is installed on a hard drive.
This genome engineering tour de force was then followed by the synthesis of a reduced Mycoplasma genome. By combing the literature and carrying out extensive mutagenesis, genes were identified that were nonessential for growth in a rich culture medium. From the design of the new genome, to its installation into a new cell, took only 3 weeks.
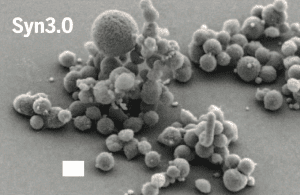
The result, Syn3.0, has 438 protein coding genes and 35 RNA genes. Its 531,000 base genome is the smallest of any autonomously replicating cell found in nature. The doubling time of the cell is 180 minutes (compared with 16 hours for M. genitalium). The cells are smaller than the parent organism and are polymorphic in apperance (illustrated; image credit).
What is encoded by this minimal cellular OS?
Most of the genes (41%) are involved in expression of the genome: transcription, regulation, RNA metabolism, translation, protein folding, RNA, ribosome biogenesis, rRNA modification, and tRNA modification.
Seven percent of the synthetic genome is involved in preservation of genome information: DNA replication, DNA repair, DNA toplogy, DNA metabolism, chromosome segregation, and cell division.
Genes involved in cell membrane synthesis constitute 18% of the genome, and genes involved in cytosol metabolism, 17%.
Perhaps the greatest surprise is that 17% of the Syn3.0 genes have no known functions. Some of these genes are also present in other organisms and must have important roles. Their study should be stimulated by the creation of Syn3.0.
I would be very excited to see this technology applied to the study of viral genomes. For most small viral genomes it has already been determined that all of the genes are needed for replication in cell culture. For example, the genome of poliovirus, a 7,500 nucleotide RNA molecule, encodes about a dozen proteins. None of these protein coding sequences can be removed without destroying the ability of the virus to replicate.
However, viruses with larger genomes carry some genes that are dispensable for replication in cell culture. For example, the DNA genomes of adenoviruses, herpesviruses, and poxviruses encode proteins that can be deleted without affecting replication in cell culture. Many of these genes encode antagonists of the immune response, and have a role only during infection of an animal with an immune system.
Undoubtedly the most interesting application of the technology used to produce Syn3.0 would come from analysis of the genomes of giant viruses such as Mimivirus, Pandoravirus, and Pithovirus. The genomes of these viruses range from 600,000 to over 2.4 million base pairs in length. They encode mostly proteins of unknown function, as well as molecules not seen in other viruses, such as components of the protein synthesis apparatus. I hope that we will soon see the synthesis of reduced genomes of these giant viruses to identify the minimal gene set needed for production of infectious viruses in a host cell.
Put another way, what is the smallest operating system needed to run a giant virus?
