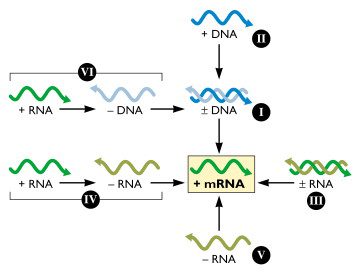

One of the most significant advances in virology of the past 30 years has been the understanding of how viral genomes are expressed. Cellular genes are encoded in dsDNA, from which mRNAs are produced to direct the synthesis of protein. Francis Crick conceptualized this flow of information as the central dogma of molecular biology:
DNA , > RNA , > protein
All viruses must direct the synthesis of mRNA to produce proteins. No viral genome encodes a complete system for translating proteins; therefore all viral protein synthesis is completely dependent upon the translational machinery of the cell. Baltimore created his virus classification scheme based on the central role of the translational machinery and the importance of viral mRNAs in programming viral protein synthesis. In this scheme, he placed mRNA in the center, and described the pathways to mRNA from DNA or RNA genomes. This arrangement highlights the obligatory relationship between the viral genome and its mRNA.
By convention, mRNA is defined as a positive (+) strand because it is the template for protein synthesis. A strand of DNA of the equivalent sequence is also called the (+) strand. RNA and DNA strands that are complementary to the (+) strand are, of course, called negative (-) strands.
When originally conceived, the Baltimore scheme encompassed six classes of viral genome, as shown in the figure. Subsequently the gapped DNA genome of hepadnaviruses (e.g. hepatitis B virus) was discovered. The genomes of these viruses comprise the seventh class. During replication, the gapped DNA genome is filled in to produce perfect duplexes, because host RNA polymerase can only produce mRNA from a fully double-stranded template.
The Baltimore classification system is an elegant molecular algorithm for virologists. The principles embodied in the scheme are extremely useful for understanding information flow of viruses with different genome configurations. When the bewildering array of viruses is classified by this system, we find fewer than 10 pathways to mRNA. By knowing only the nature of the viral genome, the basic steps that must occur to produce mRNA are readily apparent. More pragmatically, the system simplifies understanding the extraordinary life cycle of viruses.
Crick FH (1958). On protein synthesis. Symposia of the Society for Experimental Biology, 12, 138-63 PMID: 13580867
Baltimore D (1971). Expression of animal virus genomes. Bacteriological reviews, 35 (3), 235-41 PMID: 4329869

That was interesting. I never knew this method of classification existed. Not much of a virologist, but the conventional method of classification is based on whether the the virus is a DNA or RNA virus and whether the capsid has glycoproteins on it right? Correct me if I'm wrong.
I've found this site to be a nice overview of viruses showing how they are classified. It includes a picture of the virus, the genome, and a little info about how they replicate:
http://expasy.org/all_by_protein/…
Here's their page on the Hepadnaviruses mentioned above:
http://www.viralozone.expasy.org/all_by_species/…
I have a question, why there is now any virus that contain mRNA and a retro enzyme (mRNA dependent RNA polymerase) or virus that contain -DNA? it's wired that not all option are valid.
Is there no way to do phylogenetics with viruses and use phylogenetic trees instead of a classification?
Yes, phylogenetics with nucleotide sequence is now routinely used to
classify viruses. Such analyses have also been used to re-classify
viruses that were originally placed in groups by other criteria.
Yes, phylogenetics with nucleotide sequence is now routinely used to
classify viruses. Such analyses have also been used to re-classify
viruses that were originally placed in groups by other criteria.
this woz the shitiest thing i have evr herd your taking bull shitttty
Phylogenetics and other classification methods are tools. Use them wisely, whether you are trying to understand the relationships of viruses, parrots, tropical fish or gesneriads. Expect changes as relationships are better understood.
Why it doesn’t include the -ssDNA??
Pingback: Rhabdovirus genome – Negative-sense RNA (- RNA) | Medical Revision
Pingback: Rhabdovirus genome | Medical Revision
Pingback: Baltimore system of Classifications of Viruses - microbeonline