Centers for Disease Control and Prevention have determined that some adults have serum cross-reactive antibodies to the new influenza H1N1 virus. One of the techniques used to reach this conclusion is the hemagglutination inhibition (HI) assay. How does this assay work?
To understand the HI assay, we must discuss the hemagglutination assay. Influenza virus particles have an envelope protein called the hemagglutinin, or HA, which binds to sialic acid receptors on cells. The virus will also bind to erythrocytes (red blood cells), causing the formation of a lattice. This property is called hemagglutination, and is the basis of a rapid assay to determine levels of influenza virus present in a sample. To conduct the assay, two-fold serial dilutions of a virus are prepared, mixed with a specific amount of red blood cells, and added to the wells of a plastic tray. The red blood cells that are not bound by influenza virus sink to the bottom of a well and form a button. The red blood cells that are attached to virus particles form a lattice that coats the well. The assay can be performed within 30 minutes, and is therefore a quick indicator of the relative quantities of virus particles.
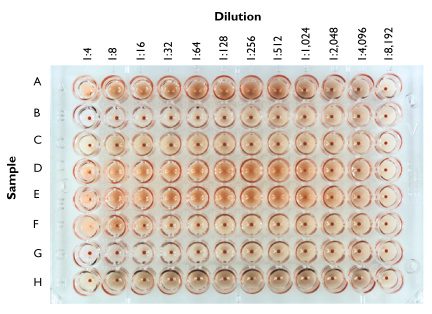

In the figure above, two-fold dilutions of samples of different influenza viruses (A – H) were prepared, mixed with chicken red blood cells, and added to the wells of a 96-well plate. After 30 minutes the wells were photographed. Sample A causes hemagglutination up to the 1:256 dilution; therefore the HA titer of this virus stock is 256. The sample in row B contains no detectable virus, while that in row D has an HA titer of 512.
The HA assay can be easily modified to determine the level of antibodies to influenza virus present in serum samples. In the CDC study cited below, the authors wished to determine whether stored serum samples contained antibodies to the new influenza H1N1 strain. First they obtained a preparation of one of the new influenza viruses, specifically A/California/04/2009 and determined its HA titer by the method described above. They added a fixed amount of virus to every well of a 96-well plate, equivalent to 32 – 64 HA units. Then they prepared two-fold dilutions of each serum to be tested, and added each dilution series along a row of wells. Finally, they added red blood cells and incubated for 30 minutes.
The basis of the HI assay is that antibodies to influenza virus will prevent attachment of the virus to red blood cells. Therefore hemagglutination is inhibited when antibodies are present. The highest dilution of serum that prevents hemagglutination is called the HI titer of the serum. If the serum contains no antibodies that react with the new H1N1 strain, then hemagglutination will be observed in all wells. Likewise, if antibodies to the virus are present, hemagglutination will not be observed until the antibodies are sufficiently diluted.
The CDC report contains the statement “…serum HI antibody titers of 40 are associated with at least a 50% reduction in risk for influenza infection or disease in populations”. A serum HI antibody titer of 40 means that at a dilution of 1:40, but not higher, the serum blocked hemagglutination. By determining HI titers and comparing them with influenza attack rates in populations, it is possible to calculate the significance of the HI antibody titer with respect to susceptibility to influenza virus infection. When used in this manner, the HI assay is a powerful epidemiological tool.
J Katz, PhD, K Hancock, PhD, V Veguilla, MPH, W Zhong, PhD, XH Lu, MD, H Sun, MD, E Butler, MPH, L Dong, MD, PhD, F Liu, MD, PhD, ZN Li, MD, PhD, J DeVos, MPH, P Gargiullo, PhD, N Cox, PhD (2009). Serum Cross-Reactive Antibody Response to a Novel Influenza A (H1N1) Virus After Vaccination with Seasonal Influenza Vaccine Morbid. Mortal. Weekly Rep., 58 (19), 521-524
Potter, CW, & Oxford, JS (1979). Determinants of immunity to influenza infection in man. Br Med Bull, 35, 69-75

In the HI-assay, one monitors surface evolution of bulk of Virus surface and needs immune antibodies to be test against new strains, to determine if the Virus surface HA protein has evolved/mutated. Are there no techniques to characterize protein evolution on single virus particles like AFM, TEM etc?
For HI test, the concentration of virus should be 8 HA unit/50 ul (4HA unit/25 ul) in each well.
Can influenza virus infect and penetrate RBCs?
Different types of blood can affect sensitivity in the assay. For example, if you are testing against seasonal flu, you would use turkey RBCs and if you are testing against avian flu, you would use horse RBCs. This is due to the binding sites on the RBCs.
The first paragraph describes the initial titration of a virus to figure out the virus titer. This plate only contains virus, PBS, and RBCs. Once the titer is determined, the virus can be back-titrated or standardized (4 HA/25ul or 8 HA/50uL) for HAI testing.
Hemp Seed Edestin protein performs this same function, and is not specific to any HxNx strain. Additionally, Gamma-Tocopherol available in Hemp Seed Oil helps to expectorate the lungs. I suggest both before bedtime, as well as integrated into one’s daily intake (flu or not).
why we keep tubes at 4 degree at haemagglutination test
I find this quite interesting and easy to comprehend… thanks for sharing…
HI Haredy,
I have the same questions. did you figure it out?
Thanks
Pingback: Prediction of antigenic phenotypes of influenza viruses | neherlab
Pingback: Pre-owned Porsche for sale
The antibodies are removed or rendered ineffective on dilution, thereby allowing the virus particle to bind with the Chicken RBC and form lattice instead of settling down the V-column and seen as a dot
Antibodies are responsible
it will but it will be delayed as the nucleus is relatively not as heavy as the chicken RBC which settles down quickly
There is not necessarily a relationship between PFU and HA titer. I work with some viruses that have a very low HA titer (20), but nearly 1×10^8 PFU/mL. Also make sure you are using an appropriate red blood cell type for the virus(es) you are working with. Dots in only the early HA dilutions can occur with viruses that have strong NA activity You may notice that hemagglutination at, say, 20 or 25 minutes turns into a dot by 40 minutes as everything warms up and the NA becomes more active (and has more time to work) and cleaves the SA off the RBC.
If haemagglutination does not occur does that suggest that that individual with that receptor on their cells, are immune to the disease?